Research
The focus of the Laboratory for Cellular Biophysics lies at the interface between physics and cell biology. Our specific interest is in the cell nucleus and establishment of biophysical imaging tools to uncover the role live cell nuclear architecture plays in genome function.
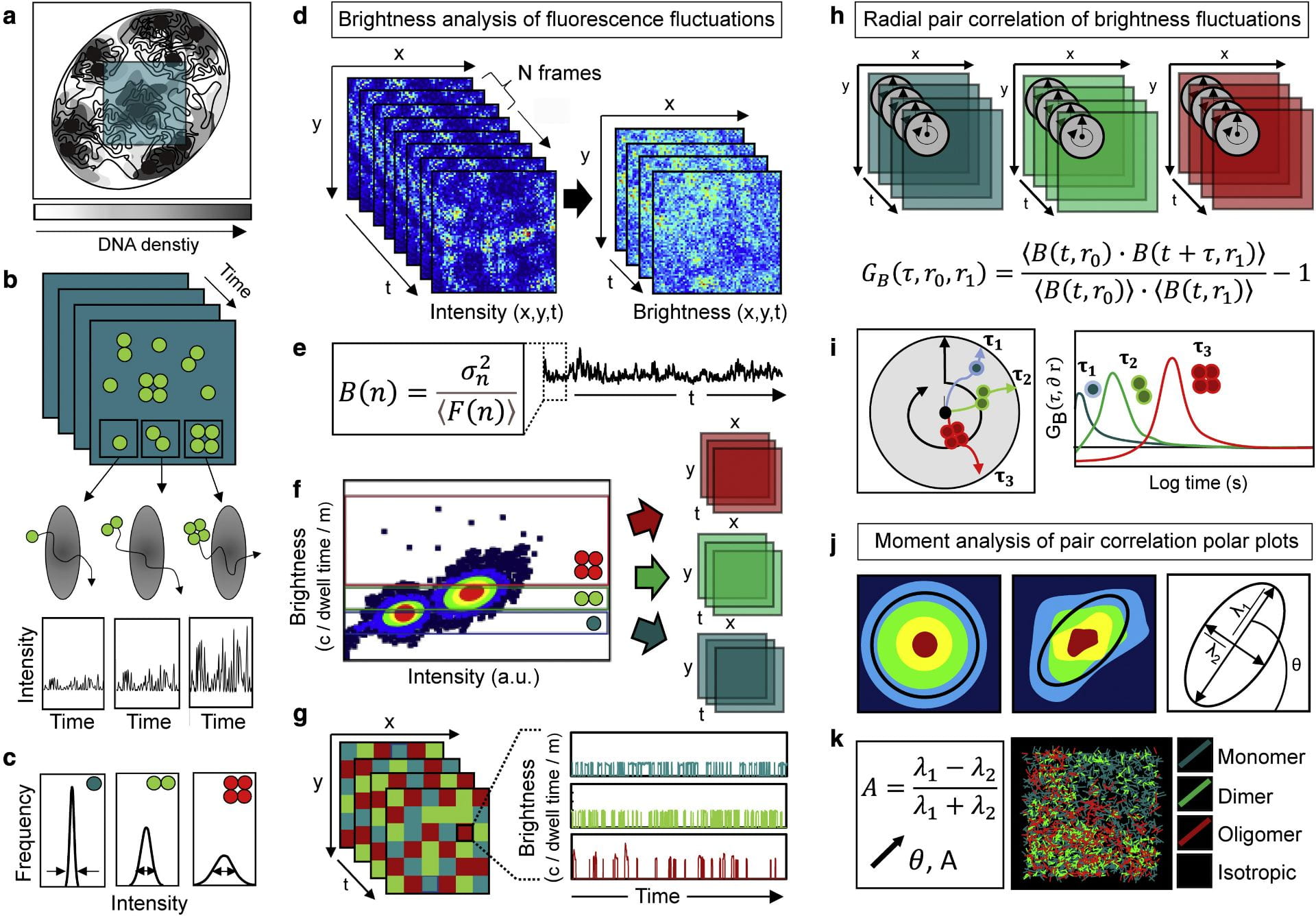
Fluorescence fluctuation spectroscopy of nuclear biophysics. We are developing fluorescence fluctuation based methods of analysis to map the diffusive route of nuclear traffic within confocal laser scanning microscopy data recording live cell fluorescent protein dynamics. This technology is based on different spatiotemporal correlation functions that have the capacity to both extract the molecular accessibility of nucleus architecture when applied to an inert fluorescent protein, and report the biophysics of DNA target search when applied to a fluorescently tagged biological protein. A particular interest in the lab is coupling spatiotemporal correlation spectroscopy with brightness based moment analysis to track nuclear protein diffusion as a function of oligomeric state. We are using this technology to quantify the impact transcription factor complex formation combined with nucleus architecture has on navigation of the nuclear landscape.
Quantitative microscopy of nucleus architecture. We are interested in understanding how the two-metres of DNA that underpins the human genome is packaged into a three-dimensional chromatin network that fits inside the microscopic volume of a cell nucleus, and how dynamic rearrangements in this structural framework, regulate DNA template access to the proteins that read, copy, and repair our genetic information. To investigate this we are spatiotemporally mapping Förster resonance energy transfer (FRET) interaction between fluorescently labelled histones core to the nucleosome (e.g., H2B) and or different architectural proteins (e.g., HP1) via use of fluorescence lifetime imaging microscopy (FLIM) and fluorescence anisotropy imaging microscopy (FAIM). Collectively this approach is enabling us to study chromatin network dynamics in a living cell on a sub-micron scale down to the level of nucleosome proximity.
Biophysical methods of analysis. Advanced fluorescence microscopy has revealed molecular insights into the spatiotemporal dynamics of biological processes that could not have been obtained by any other means. In the Laboratory for Cellular Biophysics we develop and apply methods of analysis based on fluorescence lifetime imaging microscopy (FLIM) and fluorescence fluctuation spectroscopy (FFS) to quantify protein interaction, aggregation, binding kinetics and diffusion in live cells. This includes the following:
- Fluorescence lifetime imaging microscopy (FLIM) – FRET biosensor detection
- Fluorescence correlation spectroscopy (FCS) – protein diffusion and concentration
- Moment based brightness analysis (NB, PCH) – protein oligomerisation
- Spatiotemporal correlation spectroscopy (STICS, TICS, RICS) – protein binding dynamics
- Pair correlation function analysis (pCF, pCOMB) – protein transport and accessibility